Tox21 Machine Learning Data Set
Training and test data contain both dense and sparse features
The Tox21 data set comprises 12,060 training samples and 647 test samples that represent chemical compounds. There are 801 "dense features" that represent chemical descriptors, such as molecular weight, solubility or surface area, and 272,776 "sparse features" that represent chemical substructures (ECFP10, DFS6, DFS8; stored in Matrix Market Format ). Machine learning methods can either use sparse or dense data or combine them. For each sample there are 12 binary labels that represent the outcome (active/inactive) of 12 different toxicological experiments. Note that the label matrix contains many missing values (NAs). The original data source and Tox21 challenge site is https://tripod.nih.gov/tox21/challenge/.
- Training data DENSE features: tox21_dense_train.csv.gz (51 MB)
- Training data SPARSE features:
- Training data labels: tox21_labels_train.csv.gz (<1 MB)
- Test data DENSE features: tox21_dense_test.csv.gz (1 MB)
- Test data SPARSE features:
- Test data labels: tox21_labels_test.csv.gz (< 1MB)
Code to run RandomForest:
- Python code: sampleCode.py (<1 MB)
- R code: sampleCode.R (<1 MB)
Tox21 Package
- tox21.zip(~36 MB): contains all data files and sample codes
Additional information on the data set
- Tox21 compound data in tabular format: tox21_compoundData.csv (1MB)
- Tox21 compound data in chemical format: tox21.sdf.gz (3MB)
References
If you use this data set please cite the following publications:
[Mayr2016] Mayr, A., Klambauer, G., Unterthiner, T., & Hochreiter, S. (2016). DeepTox: Toxicity Prediction using Deep Learning. Frontiers in Environmental Science, 3:80.
[Huang2016] Huang, R., Xia, M., Nguyen, D. T., Zhao, T., Sakamuru, S., Zhao, J., Shahane, S., Rossoshek, A., & Simeonov, A. (2016). Tox21Challenge to build predictive models of nuclear receptor and stress response pathways as mediated by exposure to environmental chemicals and drugs. Frontiers in Environmental Science, 3:85.
Benchmarking Results
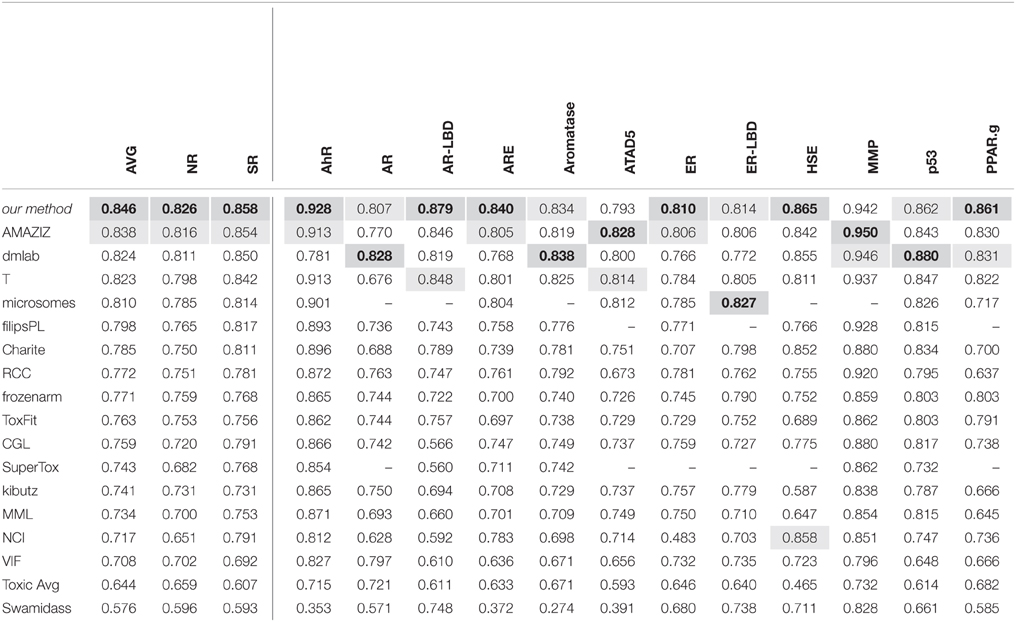 Performance of methods in terms of AUC. This table reports the performance of methods that participated in the Tox21 Data Challenge in terms of area under ROC curve. The first row, "our method", displays the performance of the Deep Learning method "DeepTox". For details see [Mayr2016]. |

