fastseg: A fast segmentation algorithm based on the cyber t-test.
"fastseg" implements a very fast and efficient segmentation algorithm.
It has similar functionality as DNACopy (Olshen and Venkatraman 2004),
but is considerably faster and more flexible. fastseg can segment data
from DNA microarrays and data from next generation sequencing for example
to detect copy number segments. Further it can segment data from RNA microarrays
like tiling arrays to identify transcripts. Most generally, it can
segment data given as a matrix or as a vector. Various data formats
can be used as input to fastseg like expression set objects for
microarrays or GRanges for sequencing data. The segmentation criterion
of fastseg is based on a statistical test in a Bayesian framework,
namely the cyber t-test (Baldi 2001). The speed-up arises from the facts,
that sampling is not necessary for fastseg and that a dynamic programming
approach is used for calculation of the segments' first and higher order moments.
This package efficiently implements the cyber t-test for segmentation of
sequential data such as intensity values for copy number detection.
The cyber t-test was proposed by P. Baldi and A.D. Long:
A Bayesian
Framework for the Analysis of Microarray Expression Data:
Regularized t-Test and Statistical Inferences of
Gene Changes , Bioinformatics, 17, 6, 509-519, (2001).
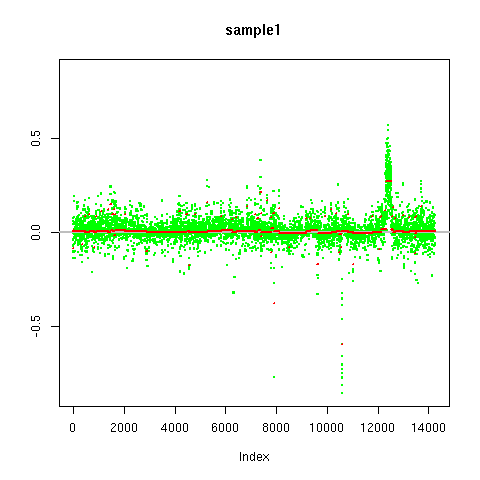 |
| Segmentation of copy number calls as produced by the cn.farms algorithm. Green dots are the copy number calls along the chromosome. On the x-axis the index of the SNP-markers are shown. The y-axis displays the copy number call. Red lines indicate the segments that were detected by fastseg. |
Paper under preparation:
"fastseg: A fast segmentation algorithm based on the cyber t-test."
Download the R-package:
- Available at Bioconductor:
fastseg R package
- To install this package, start R and enter:
source("http://www.bioconductor.org/biocLite.R")
biocLite("fastseg")

